Identification and validation of candidate clinical signatures of apolipoprotein L isoforms in hepatocellular carcinoma

[ad_1]
Demographic characteristics and mRNA expression analysis
The GSE14520 cohort contained 212 patients with HBV-related HCC. The TCGA cohort contained 370 patients with HCC. Characteristics of the cohorts are in our previous report33.
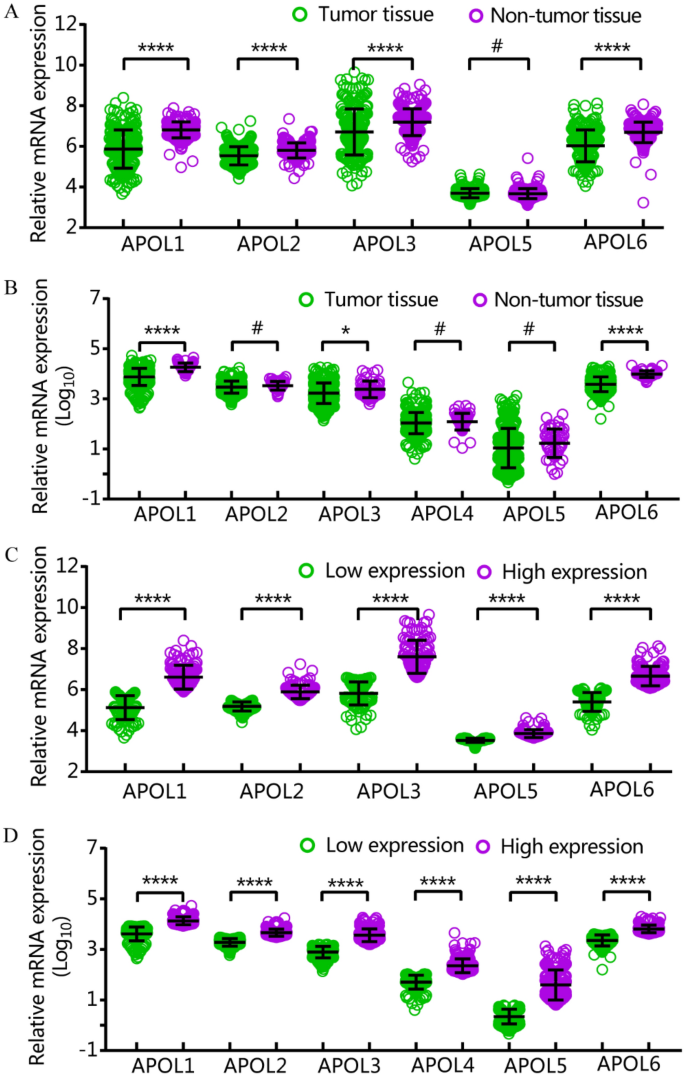
APOL1, 2, 3, and 6 were differentially expressed in tumor and non-tumor tissues in the GSE14520 cohort. APOL1, 3 and 6 were differentially expressed in tumor and non-tumor tissues in the TCGA cohort (Fig. 1A,B). All APOL isoforms were differentially expressed in low and high expression groups in both cohorts (Fig. 1C,D). APOL4 was not included in the GSE14520 cohort.
Diagnostic capacity and prognostic significance analysis
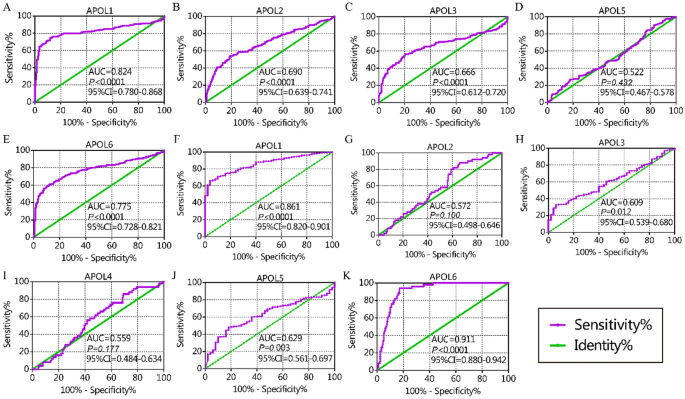
From the diagnostic capacity analysis, in the GSE14520 cohort, APOL1 and APOL6 had diagnostic significance for HCC (APOL1: area under curve [AUC] 0.824, P < 0.0001; APOL6: AUC 0.775, P < 0.0001, Fig. 2A,E). In the TCGA cohort, APOL1 and APOL6 had diagnostic significance for HCC (APOL1: AUC 0.824, P < 0.0001; APOL6: AUC 0.911, P < 0.0001, Fig. 2F,K). Others showed no or weak diagnostic capacity for HCC (Fig. 2).
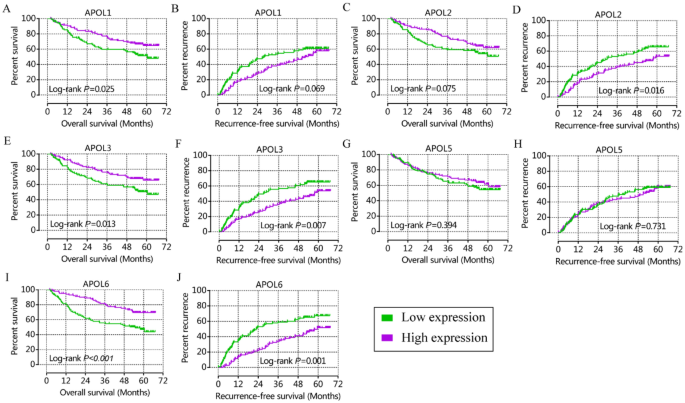
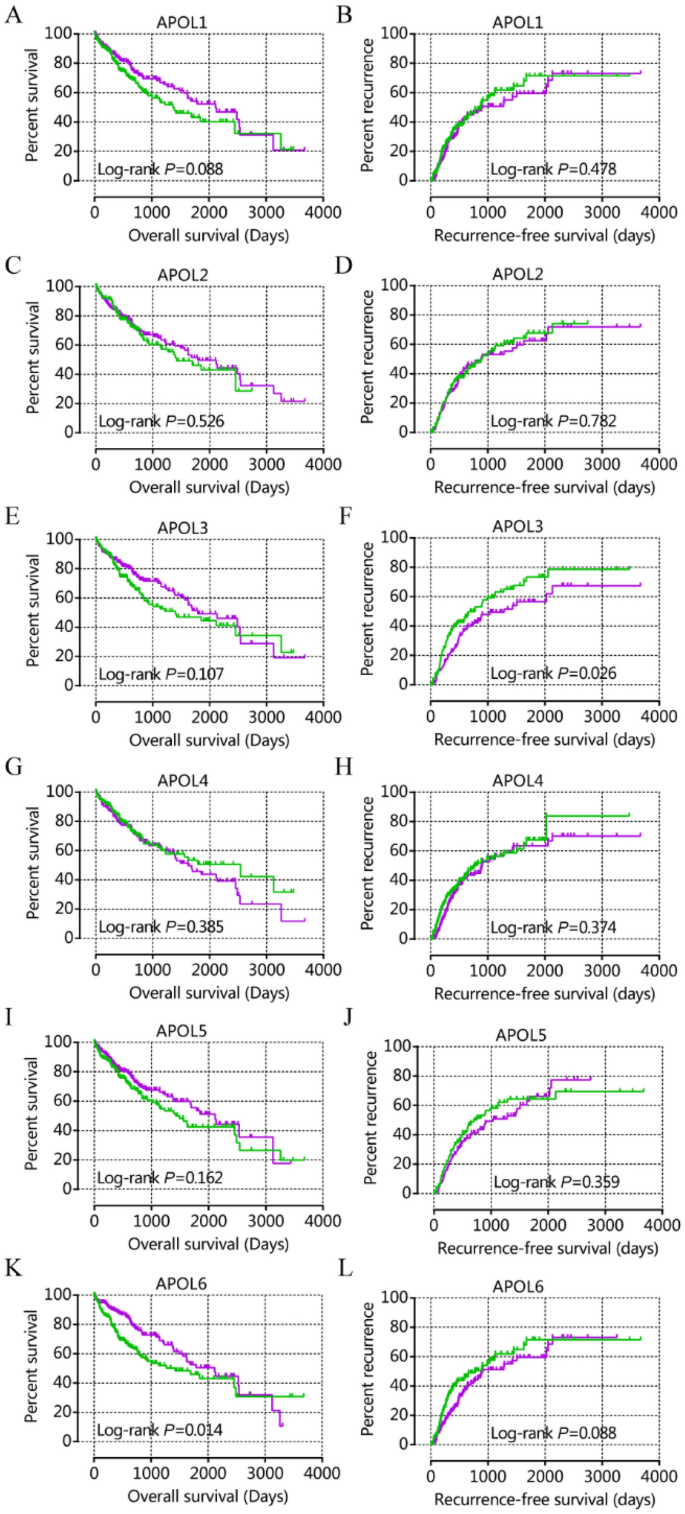
From prognostic significance analysis, in the GSE14520 cohort, APOL1, APOL3 and APOL6 showed prognostic value by univariate analysis. APOL3 and APOL6 showed prognostic value for OS by multivariate analysis (Table 1, Fig. 3). APOL2, APOL3 and APOL6 showed prognostic value for RFS in both univariate and multivariate analysis (Table 1, Fig. 3). In the TCGA cohort, only APOL6 showed prognostic value for OS in univariate and multivariate analysis (Table 2, Fig. 4). APOL3 and APOL4 had prognostic value for RFS in multivariate analysis (Table 2).
Analysis of combined prognosis-related genes
Prognosis-related genes were used for combined analysis. APOL2, APOL3, and APOL6 were combined for OS and APOL3 and APOL6 were combined for RFS in the GSE14520 cohort (Table 3, Figure S1A-E). APOL3 and APOL4 were combined for RFS in TCGA cohort (Table S1, Figure S1F). Groups containing two poor prognosis indicators had the worst survival times whereas groups with two good prognosis indicators had the best survival times.
Prospective molecular mechanism exploration by GSEA
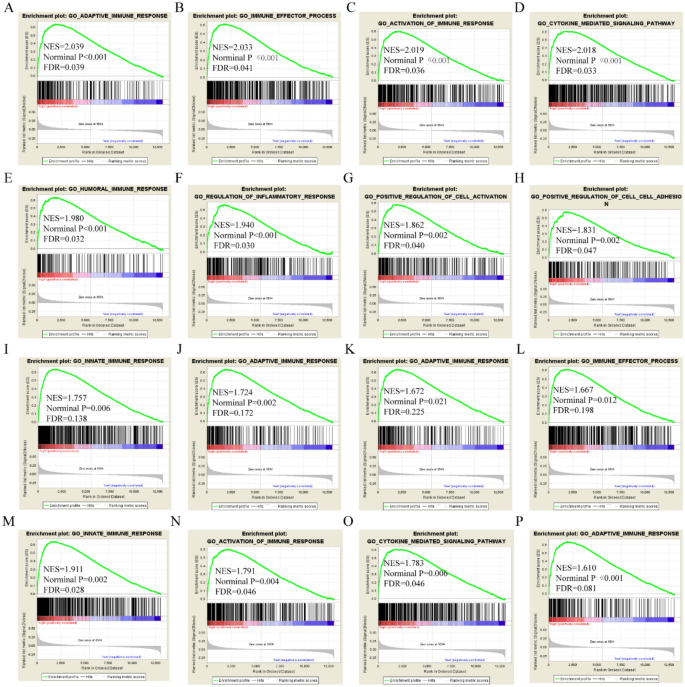
GSEA was performed to explore prospective molecular genome-wide mechanisms of APOL isoform involvement in HCC. APOL3 was involved in the adaptive immune response, immune effector processes, humoral immune response, positive cell activation, regulation of inflammatory responses, and cytokine-mediated signaling pathways by GO terms in the GSE14520 cohort (Fig. 5A–L). APOL3 was found to be involved in cell adhesion molecular cams, chemokine-signaling pathways, type 1 diabetes mellitus, and fatty acid metabolism by KEGG pathway in the GSE14520 cohort (Fig. 5M–P). APOL6 was found to be involved in the humoral immune response, fatty acid metabolism, immune effector processes, cytokine-mediated signaling pathways, and drug metabolism cytochrome P450 in the GSE14520 cohort (Figure S2).
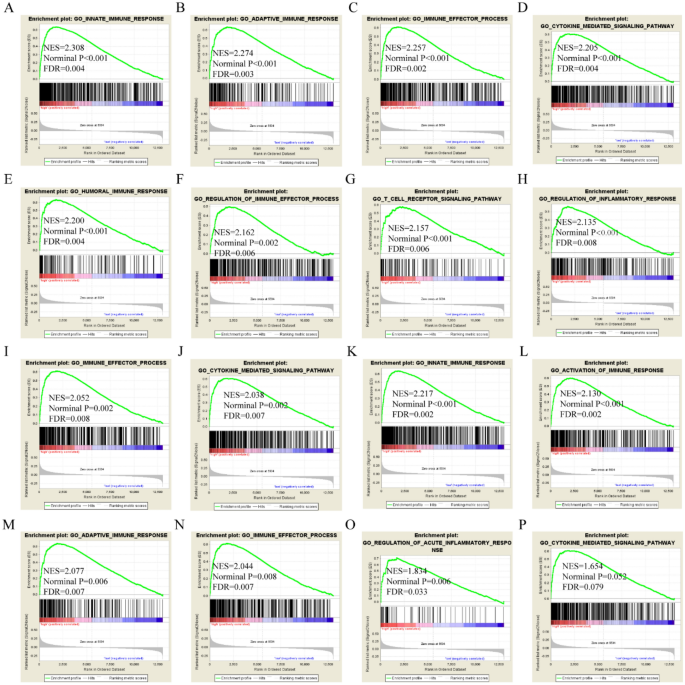
APOL3 was found to be involved in B-cell mediated immunity, activation of the immune response, the adaptive immune response, the humoral immune response, cytokine-mediated signaling pathways, chromosome centromeric region, histone binding, and chromatin binding by GO terms in the TCGA cohort (Fig. 6A–L). APOL3 was found to be involved in cell adhesion molecular cams, type 1 diabetes mellitus, chemokine signaling pathways, and fatty acid metabolism by KEGG pathways in the TCGA cohort (Fig. 6M–P). APOL6 was found to be involved in the immune effector response, B-cell mediated immunity, regulation of inflammatory responses, cytokine-mediated signaling pathways, JAK-STAT signaling pathways, and cell adhesion molecular cams in the TCGA cohort (Figure S3).
Risk score model and nomogram construction
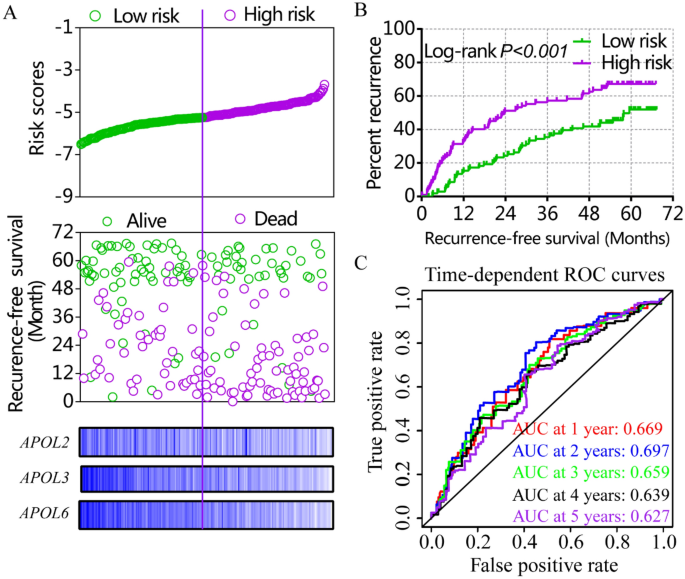
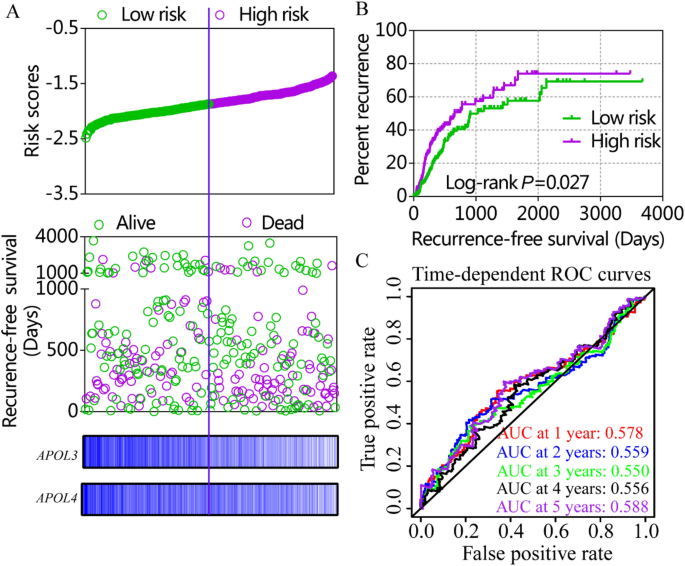
Risk score models were constructed using APOL3 and APOL6 for OS (Figure S4, Table 4) and APOL2, APOL3 and APOL6 for RFS in the GSE14520 cohort (Fig. 7, Table 4). Risk score models were constructed using APOL3 and APOL4 for RFS in the TCGA cohort (Fig. 8, Table 4). Risk score ranking, patient survival status, heat maps of APOL expression isoforms, Kaplan–Meier plots and time-dependent ROC curves for 1-, 2-, 3-, 4-, and 5-year survival were included in the models. ROC curves for the GSE14520 cohort, including OS and RFS models, showed better prognoses than for the TCGA cohort. Detailed prognostic analysis results of low- and high- risk groups were shown in Table S2.
Risk score model, Kaplan–Meier plots and time-dependent receiver operative characteristic curves for recurrence-free survival in the GSE14520 cohort. (A): Risk score model with risk score, survival status, heatmap of APOL2, 3 and 6. (B): Kaplan–Meier plots by low and high recurrence-risk groups; (C): Time-dependent receiver operative characteristic curves for recurrence-free survival at 1, 2, 3, 4, and 5 years.
Risk score model, Kaplan–Meier plots and time-dependent receiver operative characteristic curves for recurrence-free survival in the TCGA cohort. (A): Risk score model with risk score, survival status, and heatmap for APOL3 and 4. (B): Kaplan–Meier plots by low and high recurrence-risk groups. (C): Time-dependent receiver operative characteristic curves for recurrence-free survival at 1, 2, 3, 4, and 5 years.
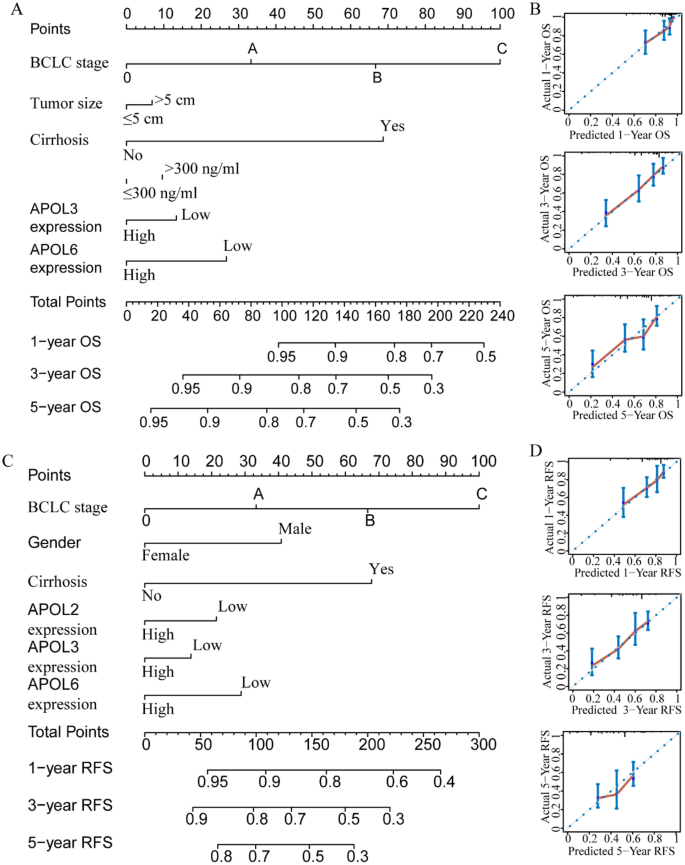
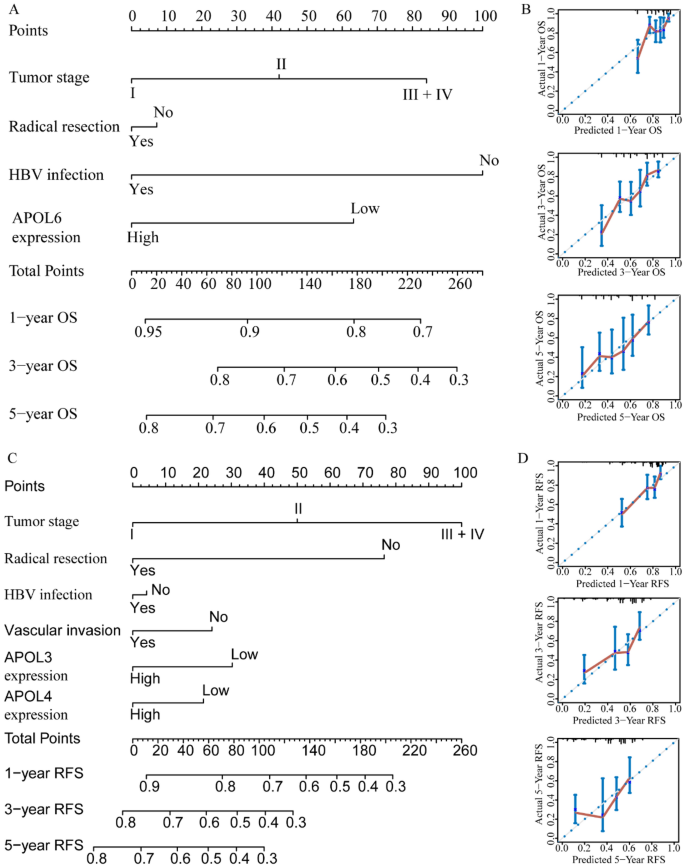
For the GSE14520 cohort, nomograms were constructed using tumor size, cirrhosis, α-fetoprotein (AFP), Barcelona Clinic Liver Cancer (BCLC) stage, APOL3 and APOL6 expressions for OS (Fig. 9A,B). Nomograms were constructed using sex, cirrhosis, BCLC stage, APOL2, APOL3 and APOL6 expression for RFS (Fig. 9C,D). For the TCGA cohort, nomograms were constructed using tumor stage, radical resection, HBV infection, and APOL6 expression for OS (Fig. 10A,B). Nomograms were constructed for tumor stage, radical resection, HBV infection, vascular invasion, APOL3 and APOL4 expression for RFS (Fig. 10C,D). Small tumor size; female sex; lack of cirrhosis; BCLC stage 0; high expression of APOL2, APOL3 and APOL6; and low AFP levels indicated higher survival rate in the GSE14520 cohort. Early tumor stage, radical resection, low APOL3 expression, high APOL4 and APOL6 expressions, vascular invasion and HBV infection indicated higher survival rates in the TCGA cohort.
Prognosis-predicted nomograms and inner validation for 1, 3, and 5 years in the GSE14520 cohort. (A-–B): Overall survival predicting nomogram using tumor size, cirrhosis, BCLC stage, AFP levels, APOL3 and APOL6 expression for 1, 3, and 5 years and inner validation for 1, 3, and 5 years. (C–D): Recurrence-free survival predicting nomogram using sex, cirrhosis, BCLC stage APOL2, APOL3 and APOL6 expression and inner validation for 1, 3, and 5 years for 1, 3, and 5 years.
Prognosis-predicting nomograms and inner validation for 1, 3, and 5 years in the TCGA cohort. (A–B): Overall survival-predicting nomogram using tumor stage, radical resection, hepatitis B virus infection status, and APOL6 expression and inner validation for 1, 3, and 5 years. (C–D): Recurrence-free survival-predicting nomogram using tumor stage, radical resection, hepatitis B virus infection status, vascular invasion, and APOL3 and APOL4 expression and inner validation for 1, 3, and 5 years.
Co-expression, protein-chemical interaction networks and matrix
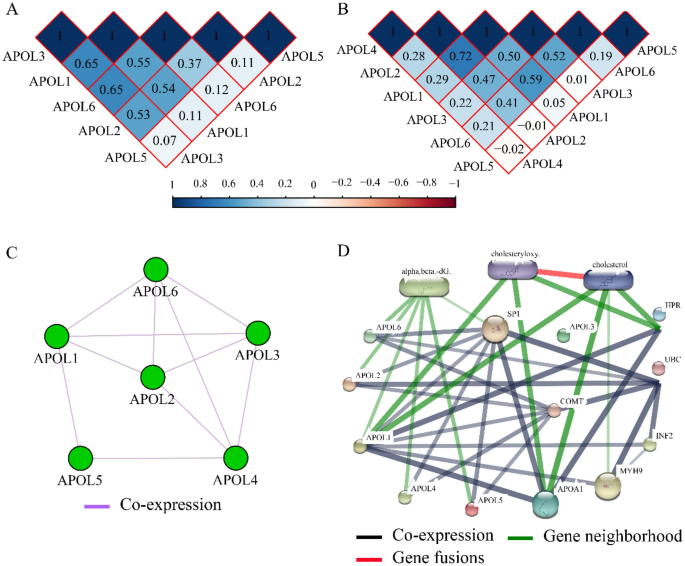
Co-expression matrixes of APOL isoforms indicated that all five APOL isoforms were positively correlated in the GSE14520 cohort. All other isoforms were positively correlated except for a negative correlation among APOL5, APOL2, and APOL4 in TCGA cohort (Fig. 11A,B). All these genes were co-expressed at the gene level (Fig. 11C). Protein-chemical interaction networks revealed that these proteins were also co-expressed at the protein level and were associated with α, β-dG, cholesteryloxy, and cholesterol in gene neighborhoods (Fig. 11D). In addition, visualized GO terms enriched by APOLs were indicated and involved in lipoprotein binding, extracellular region, lipoprotein metabolic process, et al. (Figure S5).
Co-expression matrix and protein-chemical compound interaction networks for APOL1-6. (A): Co-expression matrix for APOL1, 2, 3, 5, and 6 in the GSE14520 cohort. (B): Co-expression matrix for APOL1-6 in the TCGA cohort/ (C): Co-expression network for APOL1-6 genes. (D): Protein-chemical compound interaction networks for APOL1-6 and compounds.
Validation of diagnostic analysis and prognosis significance by Oncomine and HCCDB databases
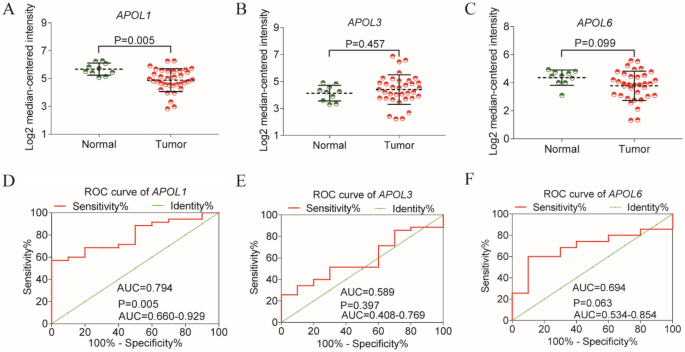
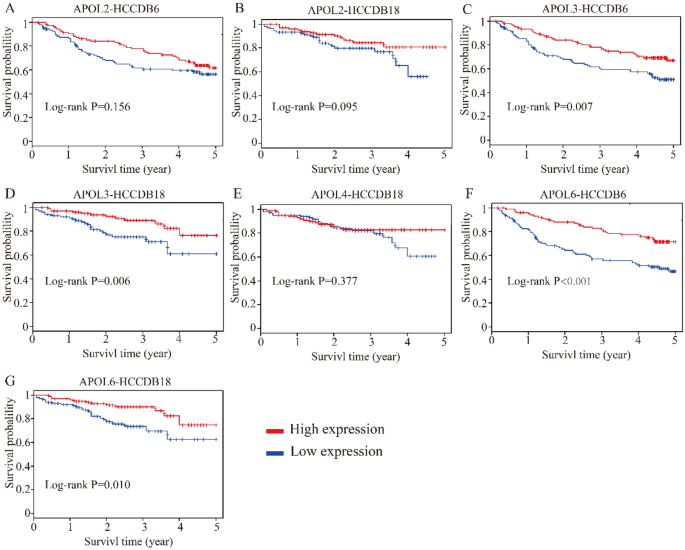
Differential expressions and diagnostic values of APOL1 and APOL3 were consistently validated in Oncomine database (AUC = 0.794, 0.589, Fig. 12A,C,D,F). Oncomine is a classic sample database in the field of cancer and can perform expression data, expression characteristics, gene set modules, etc. We applied it for validation of gene expression data. Strangely, APOL6 was showed weak diagnostic values in Oncomine database (AUC = 0.694, Fig. 12B,E). Furthermore, prognosis-related APOL isoforms were further validated in HCCDB database. HCCDB to serve as a one-stop online resource for exploring HCC gene expression with user-friendly interfaces, with integrating data from TCGA and GTEx. We applied it for validation of prognostic significance. APOL3 and APOL6, consistent in both TCGA and GSE14520 dataset, showed prognostic significance in two datasets of HCCDB as well (Log-rank P = 0.007, 0.006, Fig. 13C,D; Log-rank P < 0.001, = 0.010, Fig. 13F,G). However, APOL2 and APOL4, prognosis-related significance in TCGA or GSE14520 dataset, did not show prognostic significance (all P > 0.05, Fig. 13A,B,E).
Validation of survival analysis of APOL2, 3, 4 and 6 in HCCDB database. (A–B): Survival analysis of APOL2 in HCCDB6 and HCCDB18 datasets. (C–D): Survival analysis of APOL3 in HCCDB6 and HCCDB18 datasets. (E): Survival analysis of APOL4 in HCCDB18 dataset. (F–G): Survival analysis of APOL6 in HCCDB6 and HCCDB18 datasets.
[ad_2]
Source link